The project was initiated by the European Institute for Systems Biology and Medicine (EISBM) in collaboration with the European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI). The project team includes participants with expertise in Computational Biology, pathway curation and standard formats from research groups in France, the United Kingdom, Luxembourg, Norway, the Netherlands, Germany, the United States, Russia and Japan.
Contact
To join the effort please use this email address:
metabolismregulation@googlegroups.com
Project coordinators:
Alexander Mazein alexander.mazein@uni.lu
Irina Balaur irinaa.balaur@gmail.com
How to contribute
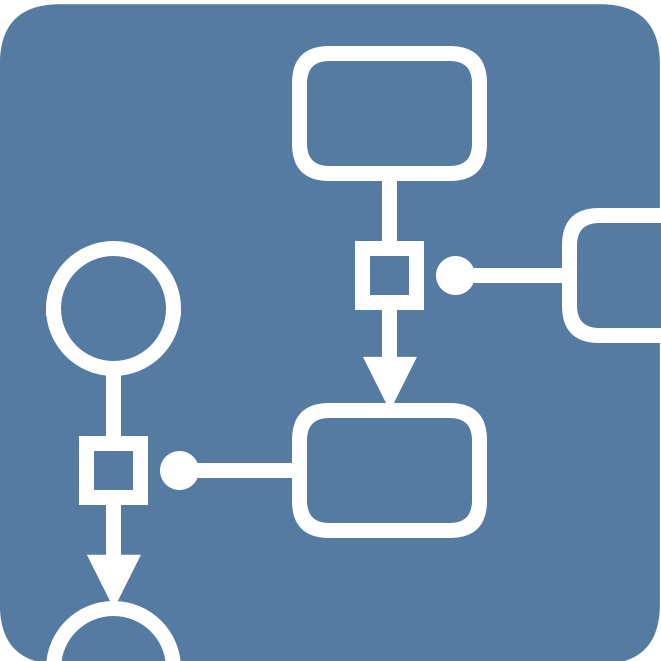
To learn more and to contribute, please review the available maps, a collection of diagrams in Systems Biology Graphical Notation (SBGN). There is a list of selected topics that are already in work or being prepared for curation.
In additon to GraphML and SBGN-ML, we aim to provide maps in various formats including CellDesigner, SBML and BioPAX, so the models are easily accessible and can be downloaded, reused and improved. These formats will be generated automatically.
Ways to contribute:
- Drawing diagrams: please join the work on one of the existing topics or propose a new one. Diagrams can be drawn using the SBGN palette in the yEd Graph Editor, the main tool in this project. Resulting format: GraphML. Please review tips on how to draw SBGN diagrams in yEd. Also, any tool that generates valid SBGN-ML can be used, for example SBGN-ED or Newt Editor, and in that case the files are to be provided in SBGN-ML.
- Proposing new topics: it can be done through direct search on the subject of metabolism regulation or by working with the automatically assembled lists.
- Proposing new colour schemes. We are looking for ways to improve the diagrams aestetically. To experiment with colours and layouts, download any diagram in GraphML format and edit as you prefer.
Project team
|
|
|
|
Irina Balaur European Institute for Systems Biology and Medicine, Lyon, France |
Alexander Mazein Luxembourg Centre for Systems Biomedicine, University of Luxembourg, Belvaux, Luxembourg |
Charles Auffray European Institute for Systems Biology and Medicine, Lyon, France |
Antonio Fabregat European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Hinxton, UK |
|
|
|
|
Adrien Rougny National Institute of Advanced Industrial Science and Technology, Tokyo, Japan |
Vasundra Touré Norwegian University of Science and Technology (NTNU), Trondheim, Norway |
Hanna Borlinghaus University of Konstanz, Konstanz, Germany |
Falk Schreiber University of Konstanz, Konstanz, Germany |
|
|
|
|
Olga Ivanova Amsterdam University Medical Centers, University of Amsterdam, Amsterdam, Netherlands |
James Greene University of Connecticut School of Medicine, Farmington, USA |
Michael Blinov University of Connecticut School of Medicine, Farmington, USA |
Tatiana Serebriiskaia Moscow Institute of Physics and Technology, Moscow, Russia |
|
|
|
|
Valeriya Berzhitskaya Moscow Institute of Physics and Technology, Moscow, Russia |
Maria Kondratova Computational Systems Biology of Cancer, Institut Curie, Paris, France |
Inna Kuperstein Computational Systems Biology of Cancer, Institut Curie, Paris, France |
Matthias König Systems Medicine of the Liver, Humboldt University of Berlin, Germany |
|
|
|
|
Marek Ostaszewski Luxembourg Centre for Systems Biomedicine, University of Luxembourg, Belvaux, Luxembourg |
Reinhard Schneider Luxembourg Centre for Systems Biomedicine, University of Luxembourg, Belvaux, Luxembourg |
Maria del Carmen University of Connecticut, Storrs, USA |
John Albanese Trinity College, Hartford, USA |
Collaborative network
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |